Acronym
REZISTOM
Contract number
J4-1769
Department:
Department of Animal Science
Chair:
Institute of Dairy Science and Probiotics
Type of project
ARIS projects
Type of project
Basic research project
Role
Lead
Financing
Duration
01.07.2019 - 30.06.2022
Total
1.45 FTE
Project manager at BF
Bogovič Matijašić BojanaABSTRACT
The intensive use of antimicrobials in human and veterinary medicine in the past 50 years has brought the problem of resistance in pathogenic microorganisms and reduced the effectiveness of infections’ treatment which the World Health Organisation declared to be one of the greatest threats to health. The fact that the commensal bacteria in food can present a reservoir of ARG have begun to draw attention of scientists years ago, but until recently there were not so advanced and powerful molecular methods and technologies available that enable the disclosure of whole bacterial genomes and microbiomes of complex samples, including the "resistome" - collection of all AR genes present in a given environment. The proposed study will be focused on the bacterial representatives of starter cultures and probiotics, which are intentionally added to the food chain. By comparative genomic analysis, we will their whole genome sequences (WGSs), we will in silico identify the ARGs, mutations and mobile elements, and determine the matching of the in silico AR predictions with the results of phenotypic detection. Furthermore, selected food samples will be examined by metagenomic sequencing in order to identify more ARGs and determine the relative abundances of these ARGs, and to asses the contribution of added starter and probiotic cultures to the resistomes of investigated food samples. The approach presented in this study, will enable efficient detection of ARGs in different resistomes associated with foods, and better assessment of the risk of transmission of ARGs along the food chain.
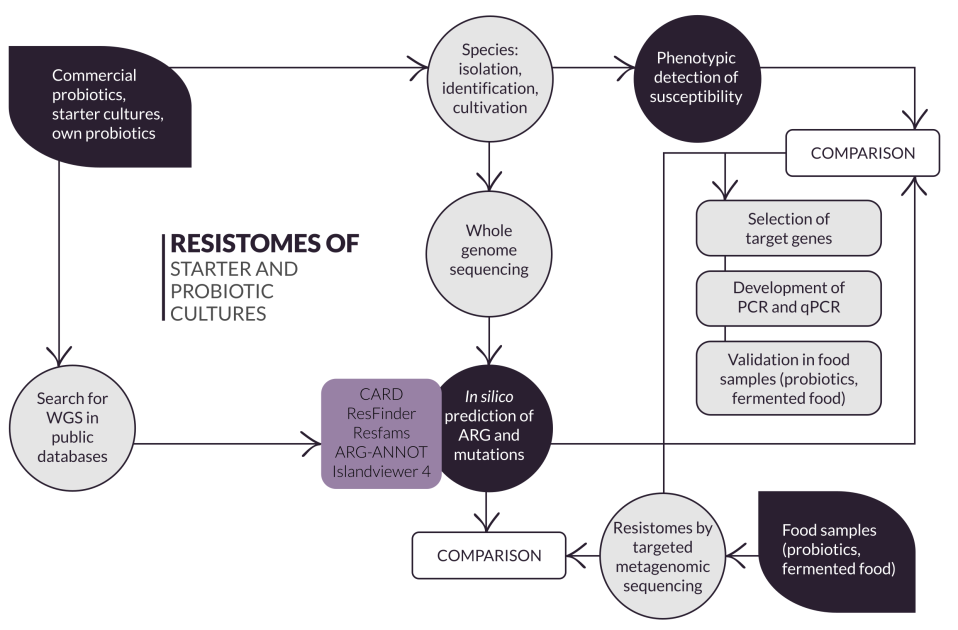
THE PHASES OF THE PROJECT AND THEIR REALIZATION
- P1: Isolation, identification and phenotypic determination of AR (Months 1-12),
- P2: Sequencing of whole genes and bioinformation analysis (Months 6-12),
- P3: In silico detection of ARGs and mutations (Months 1-24),
- P4: Development and validation of selected target ARGs with qPCR (Months 24-30),
- P5: Metagenomic sequencing of food samples and bioinformatic analysis (Months 24-32),
- P6: Synthesis of results and dissemination (writing of articles, conferences)
SELECTED PUBLICATIONS
ROZMAN, Vita, MOHAR LORBEG, Petra, TREVEN, Primož, ACCETTO, Tomaž, GOLOB, Majda, ZDOVC, Irena, BOGOVIČ MATIJAŠIĆ, Bojana. Lactic acid bacteria and bifidobacteria deliberately introduced into the agro-food chain do not significantly increase the antimicrobial resistance gene pool. Gut microbes, ISSN 1949-0984, 2022, no. 1, art. 2127438, str. 1-17, doi: 10.1080/19490976.2022.2127438.
ROZMAN, Vita, MOHAR LORBEG, Petra, ACCETTO, Tomaž, BOGOVIČ MATIJAŠIĆ, Bojana. Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. International Journal of Food Microbiology, ISSN 1879-3460. [Online ed.], 2020, vol. 314, art. 108388, str. 1-11, doi: 10.1016/j.ijfoodmicro.2019.108388.
WONG, Aloysius, BOGOVIČ MATIJAŠIĆ, Bojana, IBANA, Joyce A., LAY HONG LIM, Renee. Editorial: Antimicrobial resistance along the food chain : are we what we eat?. Frontiers in microbiology, ISSN 1664-302X, mar. 2022, vol. 13, art. 881882, str. 1-3, doi: 10.3389/fmicb.2022.881882.
LEDINA, Tijana, MOHAR LORBEG, Petra, GOLOB, Majda, ĐORĐEVIĆ, Jasna, BOGOVIČ MATIJAŠIĆ, Bojana, BULAJIĆ, Snežana. Tetracycline resistance in lactobacilli isolated from Serbian traditional raw milk cheeses. Journal of Food Science and Technology, ISSN 0022-1155, 2018, vol. 55, no. 4, str. 1426-1434, ilustr. https://link.springer.com/content/pdf/10.1007%2Fs13197-018-3057-6.pdf, doi: 10.1007/s13197-018-3057-6.
