Our research group focuses on structural and functional characterization of proteins and some other natural compounds that interact with biological and artificial lipid membranes, form transmembrane pores and/or remodel membranes, and on potential use of these proteins and natural compounds in biomedicine, pharmacy and other fields. We are using standard biochemical preparative and analytical methods and molecular biology techniques. We are preparing and characterizing model lipid forms such as artificial lipid vesicles of different sizes (SUV, LUV, GUV), lipid droplets and Langmuir lipid monolayers. We are using refractometry (surface plasmon resonance and microbalance-based surface tensiometry) to measure kinetics and interaction affinity of proteins with lipid membranes or other ligands (proteins, nucleic acids, and smaller ligands). We are characterizing enzymatic activity (especially cholinesterases and enzymes involved in oxidative stress) in biological samples and perform kinetic analyses of enzyme activity. As part of our research facilities, we also run External link to Infrastructural Centre for Molecular Interactions Analysis Open in new window.

Staff
- External link to Kristina Sepčić Open in new window, head of chair
- External link to Tom Turk Open in new window
- External link to Matej Butala Open in new window
- External link to Anastasija Panevska Open in new window
- External link to Anja Pavlin Open in new window
- External link to Gregor Bajc Open in new window
- External link to Maja Hostnik Open in new window
- External link to Luka Žeželj Open in new window
- External link to Martina Mravinec Bohte Open in new window
TEACHING ACTIVITIES
BSc and MSc programmes:
- Biology: Biochemistry, Toxinology, Bioinformatics, Genomics and proteomics, Molecular biology of membranes, Molecular biology of genes
- Microbiology: Biochemistry, Microbial biochemistry, Microbes and pathogenesis
- Biotechnology: Biochemistry, Basics of bioinformatics, Bacteriophages
- Biology-Chemistry and Biology-Home Economics (Faculty of Education): Biochemistry
PhD programmes
- Biosciences: Molecular and systemic biology, Toxins and biomembranes, The analysis of interactions between molecules using surface plasmon resonance, Natural drugs from fungi, plants and animals
- Interdisciplinary PhD study of Biomedicine: Selected topics in biochemistry and molecular biology
EQUIPMENT
The laboratory is equipped for the research in the fields of biochemistry, molecular biology and microbiology. The equipment includes:
- High-performance liquid chromatography (HPLC),
- Fast protein liquid chromatography (FPLC),
- Electrophoresis and isoelectric focusing units,
- PCR device,
- UV-VIS spectrophotometer,
- Spectrofluorimeter,
- Two VIS and fluorescence microplate readers,
- Langmuir–Blodgett trough,
- Sonicator,
- Rotavapor,
- Speed-vac,
- Two orbital shakers,
- Many centrifuges,
- Cold room.
In External link to Infrastructural Centre for molecular interactions analysis Open in new window we own two refractometers for measuring the surface plasmon resonance (Biacore X and Biacore T200), and the stopped-flow spectrometer (Stopped flow SX-20).
RESEARCH
- Biological role and applications of aegerolysin-like proteins
- Biology of bacheriophage GIL01
- Parasitic‐like modulation of Bacillus thuringiensis development and larvicidal activity by a bacteriophage
- External link to Infrastructural Centre for Molecular Interactions Analysis Open in new window
Current Research Projects
- J4-50147 Exploring the theranostic potential of aegerolysin-based protein complexes in combating bladder cancer 01.10.2023 – 30.09.2025 Kristina Sepčić
- External link to J1-4394 Parasitic‐like modulation of Bacillus thuringiensis development and larvicidal activity by a bacteriophage Open in new window1.10.2022 - 30.09.2022 Butala Matej
Current Research Programs
- External link to P1-0207 Toxins and biomembranes Open in new window (head of program: prof. dr. Igor Križaj, funded by ARRS)
Aegerolysins: Biological role and applications
Aegerolysins are ~15 kDa proteins that interact with specific membrane lipids, and are produced mainly by bacteria and fungi. Selected aegerolysins from the mushroom genus Pleurotus recognize, and bind to organism-specific sphingolipids; therefore, they can be used as universal markers of these lipids in living cells and in artificial lipid systems. In the presence of their protein partner bearing the membrane-attack complex/perforin (MACPF) domain, aegerolysins form transmembrane pores in target membranes, causing an immediate death of the cell. Cytolytic complexes based on aegerolysins and their MACPF-protein partners can be used for selective elimination of cells harbouring the aegerolysin lipid target in their membranes. In our group, we have been studying aegerolysin-based cytolytic complexes that exert selective toxicity towards coleopteran insect pests, and complexes that are selectively toxic for urothelial cancer cells (Figure).
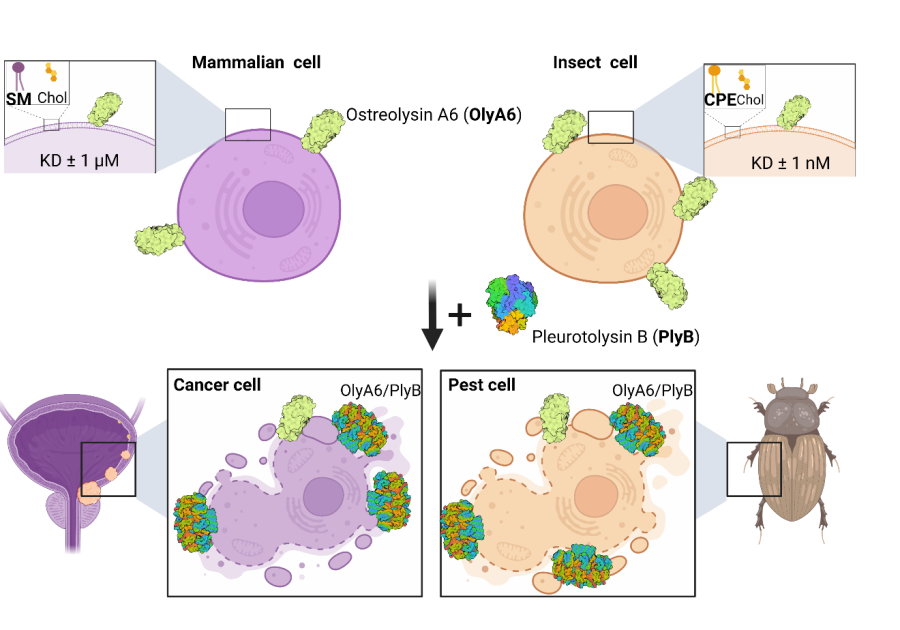
Applications of cytolytic complexes based on an aegerolysin protein OlyA6. Left: Mammalian cells. OlyA6 recognizes a specific cholesterol-bound conformation of sphingomyelin (SM) in lipid membranes. In addition, in the presence of the MACPF-domain protein partner pleurotolysin B (PlyB), OlyA6 forms transmembrane pore complexes that kill the target cells. Urothelial carcinoma cells are enriched in SM/cholesterol domains and represent a potential target for their elimination by the OlyA6/PlyB complex [Resnik in sod., 2015; PLoS ONE 10(9): e0137878]. Right: Insect cells. OlyA6 binds with 1000-fold higher affinity to the invertebrate-specific membrane sphingolipid ceramide phosphoethanolamine (CPE) than to membrane SM. The combination of OlyA6 with PlyB can lyse pest cells expressing CPE, and thus these complexes can be used as novel bioinsecticides for pest control [Panevska in sod., 2019; Sci Rep 25;9(1):5073].
Find more External link to here Open in new window.
Biology of bacteriophage GIL01
The GIL01 bacteriophage, is a tectiviral temperate phage, which infects the insect pathogen Bacillus thuringiensis. Unlike most temperate phages, GIL01 lysogeny is not established by a dedicated phage repressor, but rather by the host’s regulator of the SOS response, LexA. However, LexA is unable to maintain lysogeny, unless the small phage-encoded protein gp7 is also present. Gp7 directly interacts with LexA to enhance its DNA binding to phage promoter. We obtained the crystal structure of the 50-amino acid gp7 protein and according to SAXS data, we generated a structural model of gp7 in complex with LexA, which illustrates that gp7 positions LexA in a DNA bound conformation. By applying surface plasmon resonance approach we also recently discovered, that gp7 homologs identified in other tectiviruses, that infect a diverse range of bacteria, exhibit similar mode of action as gp7. Our recent results furthermore show, that a small protein non-homologous to gp7 in an important human pathogen, analogously to gp7, interacts with its cognate DNA damage response repressor to modulate the SOS response. Thus this results lead us to think that this kind of mechanism is widely spread to enable the SOS control beyond the LexA/RecA regulation. To determine how GIL01 establishes the lytic cycle, we examined the regulatory mechanisms at the lytic promoter. We show that lytic promoter is also repressed by LexA/gp7 complex and that the second phage-borne small protein, gp6, is the key activator of the lytic cycle. Surprisingly, gp6 is homologous to LexA itself and, thus, is a rare example of a LexA homologue directly activating transcription. We propose that the interplay between these two LexA family members, with opposing functions, ensures the timely expression of GIL01 phage late genes.
Find more External link to here Open in new window.
Bacteria are preyed upon by bacteriophages (phages), which, like other viruses, hijack the host`s cellular machinery to replicate. It is becoming increasingly clear that phages can have a significant impact not only on bacteria, but also on eukaryotes. In this project, we will determine the physiological changes in the bacterium Bacillus thuringiensis serovar israelensis (including sporulation rate and production of crystal toxins) and insect host enforced by phage-encoded proteins. It is worth noting that majority of biolarvicidal products are based on entomopathogenic B. thuringiensis serovar israelensis. In addition, we will test whether the phage genome (prophage) is packaged in endospores that can allow safe entry of prophage into the insect body. In addition, we will investigate whether phage regulators that affect processes in bacteria, also affect insect larvae. We will also investigate whether similar regulators that modulate phage-baterium-larvae interaction, exist also in other phage or bacteria.
We believe that the findings from this project will provide new fundamental knowledge to understand how phage and bacterial transmission is facilitated to other trophic levels.
Mosquitoes are one of the most recognized diseasse carrying vectors. The results of this project could pave the way for the development of improved biopesticides.
RESEARCH ARCHIVE
- External link to Actinoporins - α-pore forming toxins Open in new window
- External link to Characterization of pharmacologically active natural products from marine organisms Open in new window
- External link to Nanoemulsions and artificial lipid droplets Open in new window
- External link to Construction of a method for identification of directly or indirectly bound proteins at specific loci on bacterial chromosomes Open in new window
More research papers by members can be found External link to here Open in new window.
