Lactic acid bacteria (LAB) and bifidobacteria can be a reservoir of antimicrobial resistance, but the risk posed by strains deliberately introduced into the food chain has not been thoroughly investigated. The objective of this study was to determine whether probiotics, starter and protective cultures, and feed additives pose a risk to human health due to the possible presence of transmissible genes for antibiotic resistance. In addition to commercially available strains of lactic acid bacteria and bifidobacteria, isolates from breast milk, colostrum, human intestinal mucosa, human feces, and fermented foods were included. Phenotypic susceptibility data from 474 strains showed that antimicrobial resistance was more common in isolates from the gut than in commercial strains (Figure 1). Using comparative genomics, antimicrobial resistance genes (ARGs) and mobile genetic elements (MGEs) were found in the whole genome sequences of 1114 strains. Intrinsic ARGs were abundant in enterococci, bifidobacteria, and lactococci, but due to the absence of MGEs, we do not consider them risky. The results showed that 13.8% of commercial strains contained acquired ARGs, most frequently those for tetracycline. 75.5% of acquired ARGs were associated with known or novel MGEs, and their transmission potential was assessed based on a review of metagenomic sequences available in public collections. We confirmed that commercial strains had fewer ARGs and MGEs and were not as diverse as those found in human gut isolates or breast milk isolates, suggesting that strains intentionally introduced into the agricultural food chain do not pose a greater risk. Nevertheless, it is reasonable to pay more attention to individual probiotic strains that contain elements that have been shown to have a high potential for transfer to the gut microbiota.
In silico analyzes of whole genomes (presence of antibiotic resistance genes, mobile genetic elements, and mutations), bioinformatic analyzes of pangenomes and nuclear genomes, and analyzes of metagenomic data were largely performed by then young researcher External link to dr. Vita Rozman Open in new window with the help of External link to Asst. prof. dr. Tomaža Accetto Open in new window. Asst. dr. Majda Golob and Assoc. prof. dr. Irena Zdovc from the Veterinary Faculty were mainly involved in phenotypic determination of antibiotic resistance. External link to Asst. dr. Petra Mohar Lorbeg Open in new window and External link to Asst. prof. dr. Primož Treven Open in new window participated in obtaining and examining samples and isolates, phenotypic characterization, and writing the paper. External link to Sen res. fell. dr. Bojana Bogovič Matijašić Open in new window, head of the J4-1769 project and J4-0097 program, led the research and supervised the young researcher Vito Rozman as a PhD supervisor and participated in the writing of the article P4-0097.
The research findings presented in the article contribute significantly to more effective identification and assessment of the risk of antibiotic resistance transmission along the food chain associated with the use of bacterial strains intentionally introduced into the food chain.
The research was conducted as part of ARRS project J4-1769 Resistomes of probiotic and starter cultures as a potential risk factor for the spread of antibiotic resistance, program P4-0097 Nutrition and microbial ecology of the gastrointestinal tract, and the PhD training of young researcher Vita Rozman, who completed her PhD studies in Biosciences at the Biotehnical Faculty, University of Ljubljana.
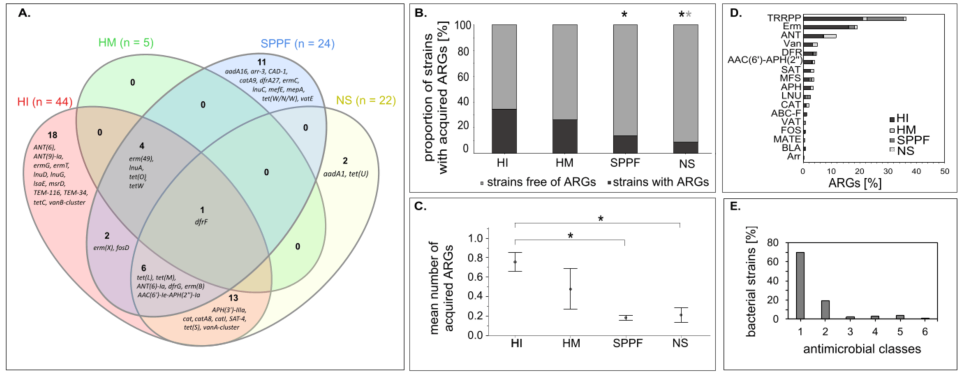
Figure 1. Acquired antimicrobial resistance genes (ARGs) in lactic acid bacteria and bifidobacteria of different origin. (A) Venn diagram showing acquired ARGs in four groups of bacteria. (B) The proportion of strains with acquired ARGs. Statistics: Likelihood ratio test, Fisher's exact probability test; p < 0.05; black asterisk, statistically significant difference compared to the human intestinal mucosa or faeces group (HI); grey asterisk, statistically significant difference compared to the human milk or colostrum group (HM). (C) The mean number (± standard error) of acquired ARGs. Statistics: rank analysis of variance (Kruskall-Wallis), pairwise multiple comparisons using Dunn’s test with Bonferroni adjustment; p < 0.05; *, statistically significant difference. (D) The prevalence of the protein families of acquired ARGs. (E) The prevalence of the strains according to the number of antimicrobial classes towards which the acquired ARGs confer resistance. NS, isolates of the natural microbiota from fermented products (non-starter strains); SPPF, strains intentionally introduced into the agro-food chain via starter, protective, and probiotic cultures and feed additives; AAC(6')-APH(2''), aminoglycoside acetyltransferase/phosphotransferase; ABC-F, ABC-F subfamily of ATP-binding cassette proteins; ANT, aminoglycoside nucleotidyltransferase; APH, aminoglycoside phosphotransferase; Arr, rifampin ADP-ribosyltransferase; BLA, β-lactamase; CAT, chloramphenicol acetyltransferase; DFR, dihydrofolate reductase; Erm, Erm 23S rRNA methyltransferase; FOS, fosfomycin thiol transferase; LNU, lincosamide nucleotidyltransferase; MATE, multidrug and toxic compound extrusion efflux pump; MFS, major facilitator superfamily efflux pump; SAT, streptothricin acetyltransferase; TRRPP, tetracycline-resistant ribosomal protection protein; Van, glycopeptide resistance gene cluster; VAT, streptogramin acetyltransferase.
Source:
ROZMAN, Vita, MOHAR LORBEG, Petra, TREVEN, Primož, ACCETTO, Tomaž, GOLOB, Majda, ZDOVC, Irena, BOGOVIČ MATIJAŠIĆ, Bojana. Lactic acid bacteria and bifidobacteria deliberately introduced into the agro-food chain do not significantly increase the antimicrobial resistance gene pool. Gut microbes. 2022, no. 1, art. 2127438, str. 1-17, ilustr. ISSN 1949-0984. External link to https://www.tandfonline.com/doi/full/10.1080/19490976.2022.2127438 Open in new window, DOI: External link to 10.1080/19490976.2022.2127438 Open in new window. [COBISS.SI-ID External link to 123511555 Open in new window]
Founding:
ARRS [J4-1769, P4-0097, funding of a young researcher Vita Rozman 6316-1/2017-273, 603-1/2017-13]